-
A vintage year for African malaria vector genomics
Scientific highlights from 2024.
-
A coding assistant for genomic data analysis?
AI coding assistants are getting very good. Would it be possible to create an AI assistant to support generation of code to analyse genomic data on malaria mosquitoes?
-
Faster neighbour-joining
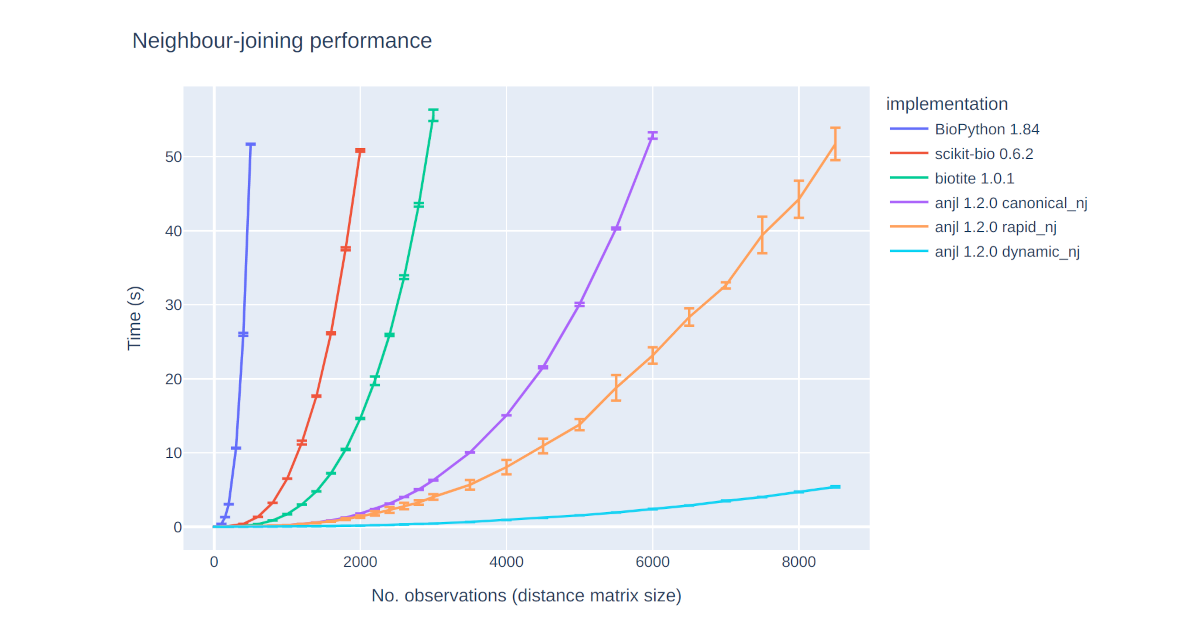
A new Python package called anjl provides fast implementations of the neighbour-joining algorithm and interactive plotting of unrooted trees.
-
Anopheles coluzzii in Kenya - neighbour-joining tree analysis
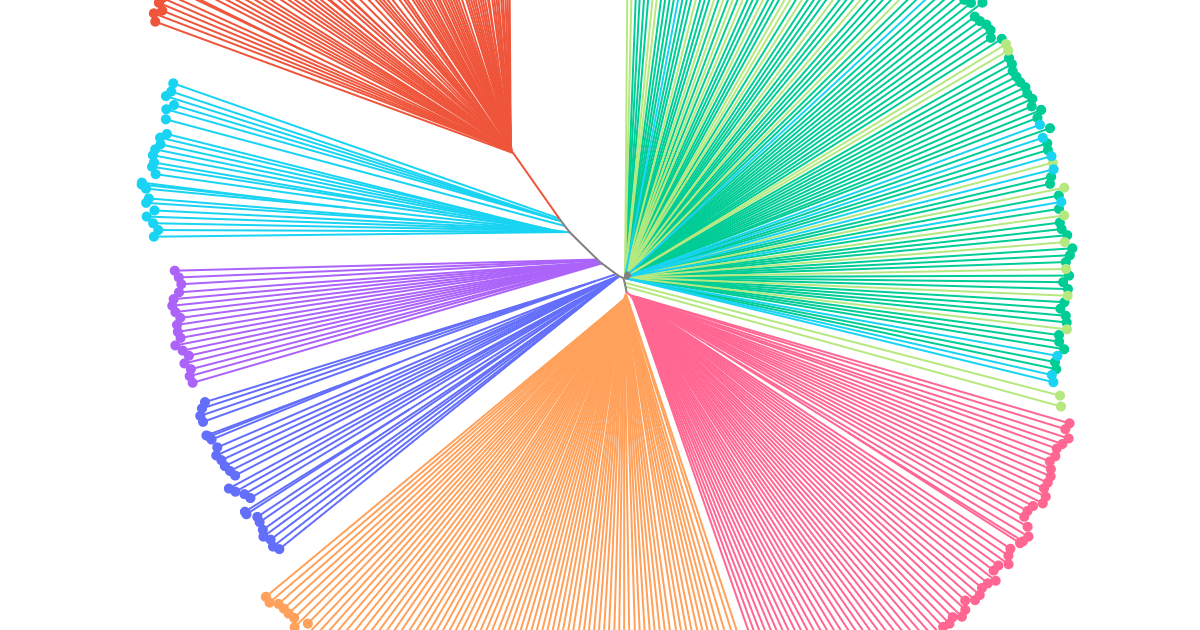
Replicates the neighbour-joining tree analysis from Kamau et al. (2024) on Anopheles coluzzii in northwestern Kenya and demonstrates the plot_njt() function in the malariagen_data Python package.
-
Blogging with Jekyll, Jupyter notebooks and interactive plots (Plotly, Bokeh)
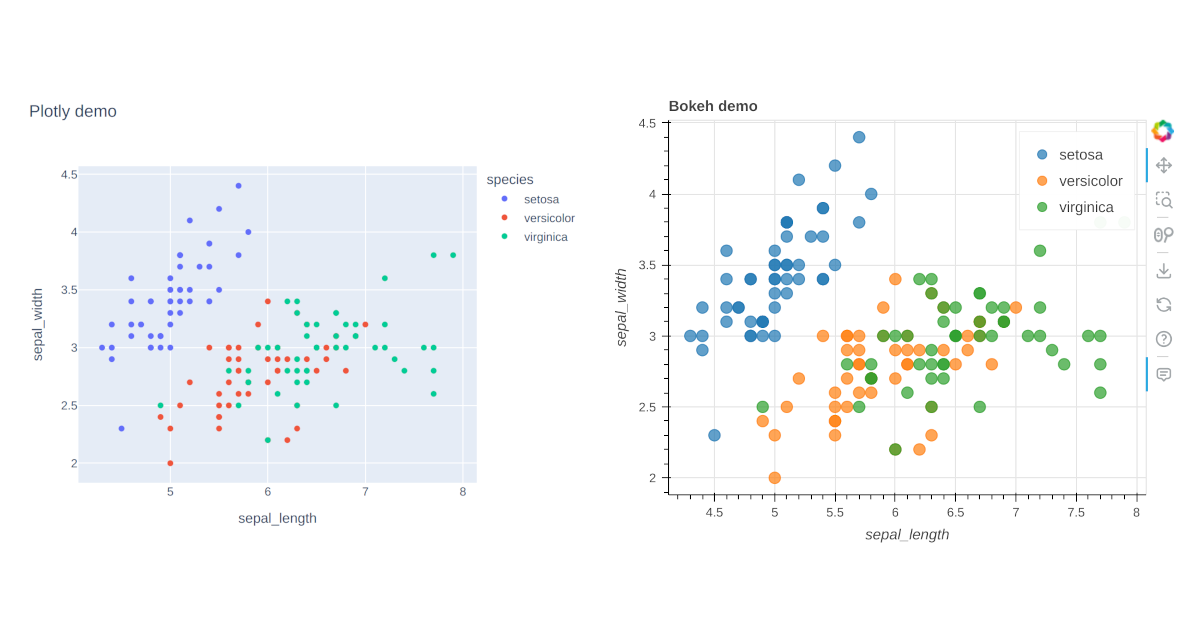
How to create blog posts from Jupyter notebooks which include interactive plots made with Plotly or Bokeh.
-
Diplotype clustering
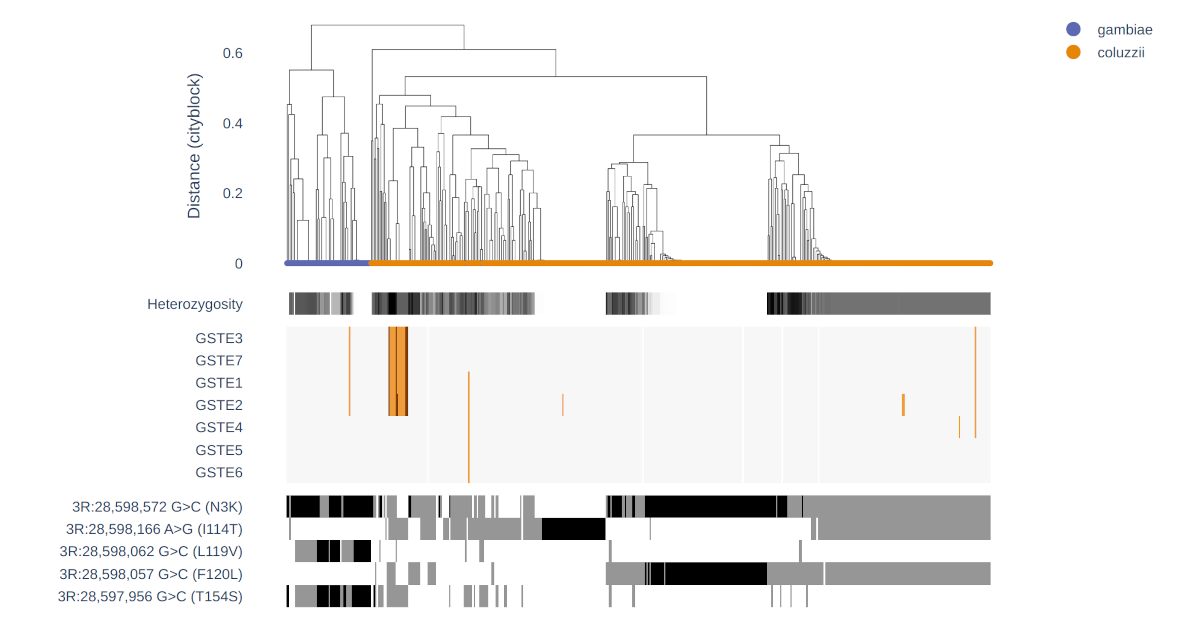
Introduces advanced diplotype clustering, a new function in the malariagen_data Python package for investigating insecticide resistance mutations.
-
Arthropod Genomics Symposium 2024

Highlights from the Arthropod Genomics Symposium in Nairobi - gene drives, microbiome, genome assemblies and insecticide resistance.
-
The Malaria Vector Genome Observatory is online
Introducing a new web page for the Malaria Vector Genome Observatory providing an entry point to data, training, software, research and more.
-
20,000 Anopheles mosquito genomes

The Malaria Vector Genome Observatory now contains analysis-ready data from whole-genome sequencing of 22,015 Anopheles mosquitoes.
-
Training course in data analysis for genomic surveillance of African malaria vectors

This post introduces a new training course and explains some of the rationale and technology choices we made to create, publish and deliver the course online.
-
Kigali, September 2022

Reflections from the PAMCA Conference 2022 in Kigali, Rwanda.
-
Trust me, I'm a doctor!

6 years, 2 babies and a pandemic later, I finally graduated as a Doctor of Philosophy. It was a sunny, winter's day in Oxford, and I got to share the experience with my wife and eldest daughter, it was a real treat. Here is a brief overview of my thesis and a few personal reflections on doing a PhD later in life.
-
Ag1000G phase 3 lightning talk at GEM 2021

A brief introduction to the open genomic data resources on malaria mosquitoes being created in phase 3 of the Anopheles gambiae 1000 Genomes Project.
-
Dask thought: distributed clusters and worker memory/storage
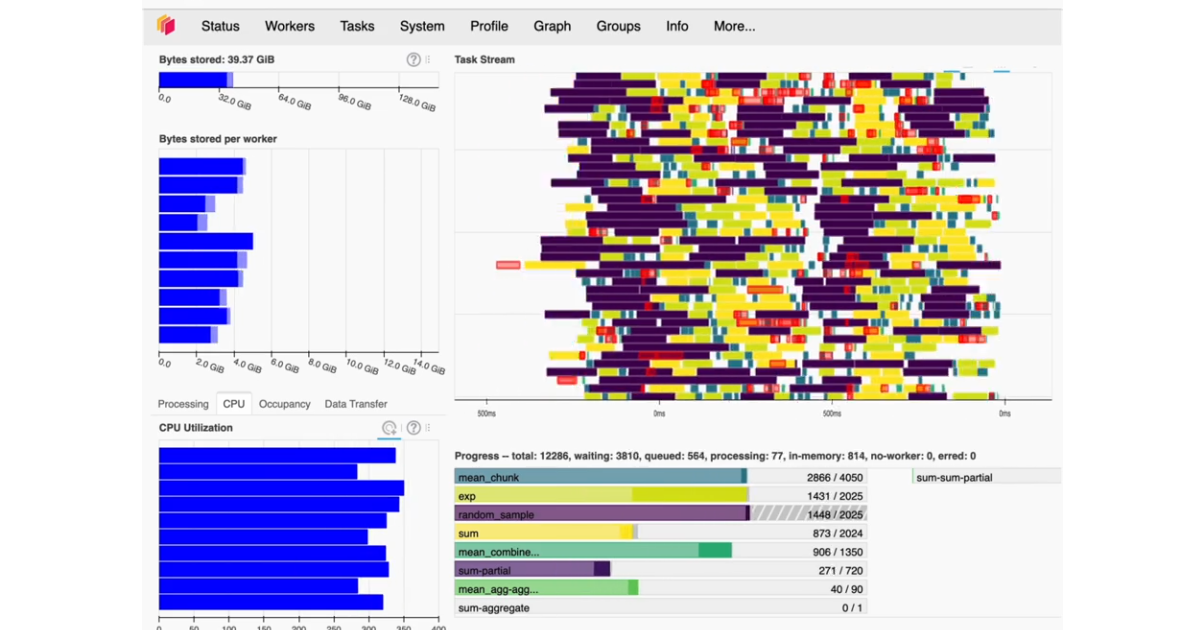
Some thoughts about Dask clusters and memory management.
-
Querying multidimensional data with xarray
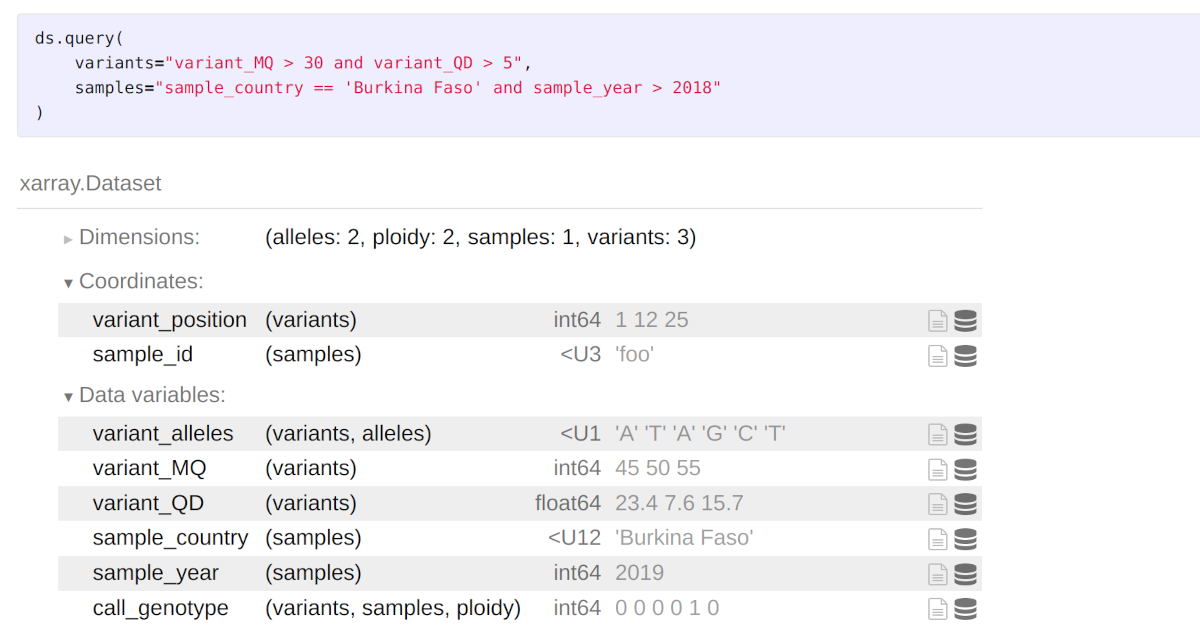
A new query() method for the xarray Dataset class.
-
On the evolution of pyrethroid target-site resistance
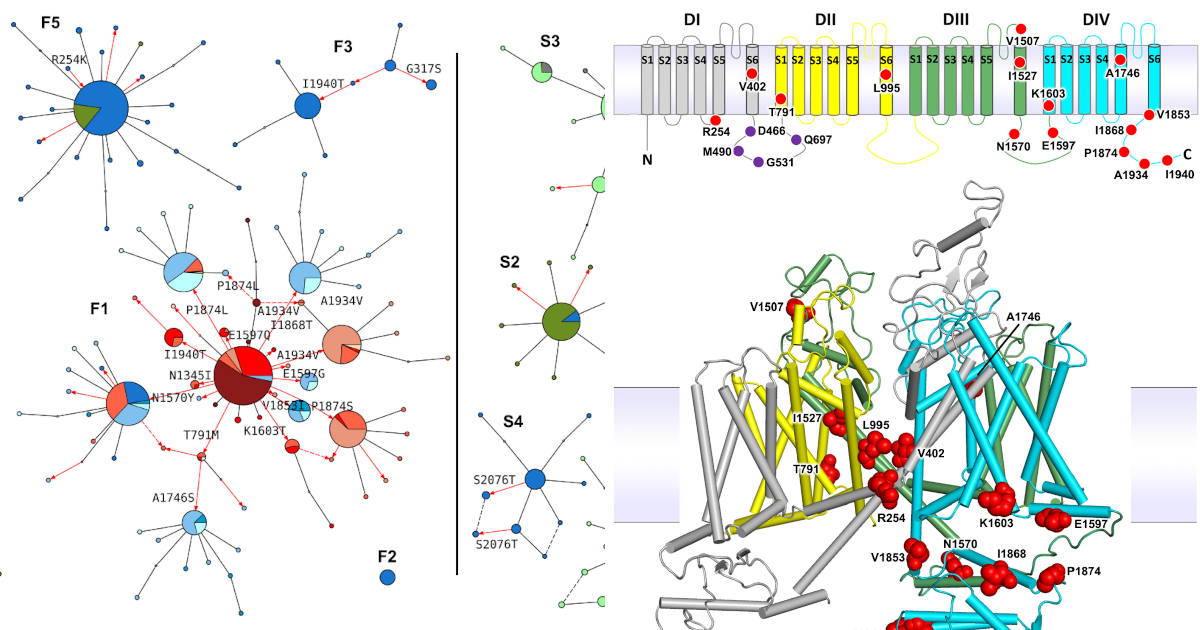
There are multiple double- and triple-mutant combinations in the Vgsc gene which I think we should be more concerned about, and they are evolving in different ways.
-
Insecticide resistance gene copy number variation in malaria mosquitoes
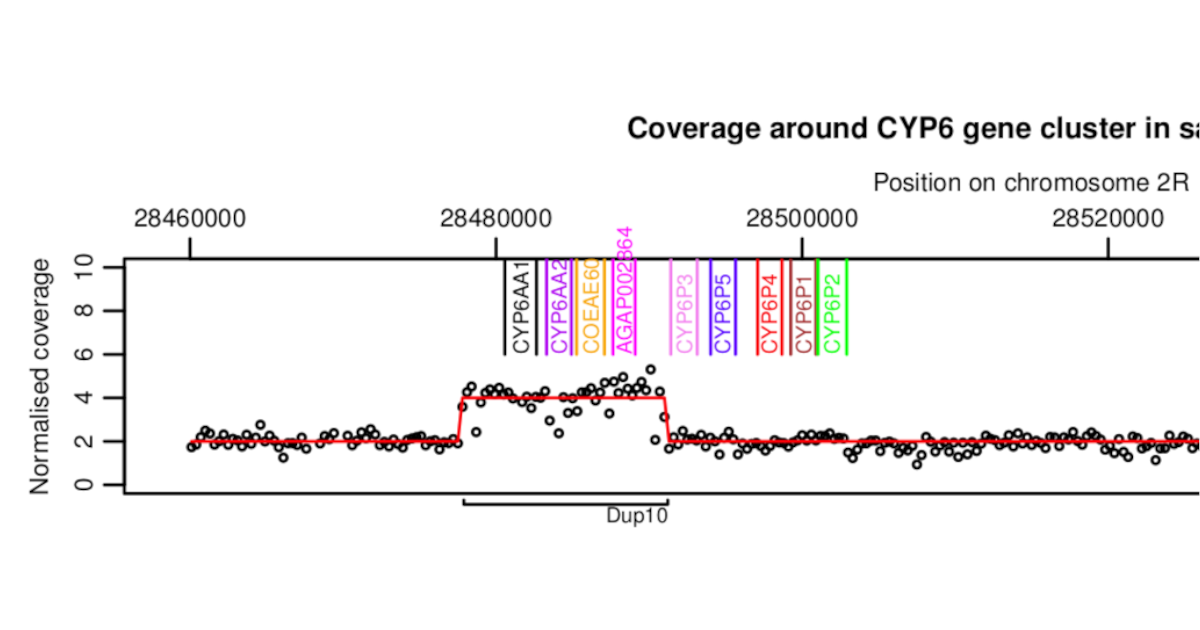
Introduces recent work on copy number variation by the Anopheles gambiae 1000 Genomes Project.
-
Zarr protocol v3 design update

Shares some slides summarising the current state of exploratory work on the Zarr v3 protocol spec.
-
Zarr Python 2.3 release
-
Senior post-doc in malaria vector genomics and modelling
-
Zarr 2.2 tech dive
-
Selecting variants and samples
-
Web-based exploration of genome variation data
-
Extracting data from VCF files
-
Installing Python for data analysis
-
Mendelian transmission
-
VGSC gene models
-
Go faster Python
-
Plotting African ecosystems
-
Genotype compressor benchmark
-
Zarr 2 - groups, filters and Zstandard
-
Understanding PBWT
-
A tour of scikit-allel
-
XKCD-style plots
-
CPU blues
-
To HDF5 and beyond
-
Mosquito genomes and malaria control, the podcast
-
Genetic variation in parasite crosses
-
Fast PCA
-
Estimating FST
-
Introducing scikit-allel
-
Blog, migrated
subscribe via RSS