Arthropod Genomics Symposium 2024

I felt very lucky to attend the Arthropod Genomics Symposium in Nairobi last week. The scientific talks were excellent, and there aren’t many other meetings where you get the same crossover of folks working on disease vectors, agricultural pests and species in need of conservation - three different fields of applied genomics but lots of common challenges and new ideas. The last AGS I attended was in 2017 and so it was great to reconnect and see how much things have progressed.
I presented on accelerating genomic surveillance of malaria vectors in Africa. More than 23,000 African Anopheles genomes have now been sequenced through concerted efforts by MalariaGEN, PAMCA, GAARD, Target Malaria, PAMGEN and many others. The main question is, wow can we now build on this to address the challenges of resistance to dual AI LLINs and other evolutionary threats to vector control? My slides are here.
I also compiled some highlights below from other talks with links to papers where I could find them.
Drew Hammond (@DrewMarcHammond) showed that viable resistance to gene drives in Anopheles is possible even in ultra-conserved regions like doublesex. Resistance can be mitigated by multiplexing targets though. Paper coming soon from Mariaou et al.
Jonathan Kayondo gave an update on the @TargetMalaria project developing gene drives for Anopheles, and work happening on non gene drive male bias at @UVRIug in Uganda.
George Christophides (@ChristophidesG) presented transmission-blocking gene drives for malaria in the Transmission Zero project (@transm0) and work in Africa to develop and test a two-component gene drive system. See Hoermann et al. 2022.
Zhijian Jake Tu (@Zhijian_Tu) shared work on the sex determination system in Aedes. Female mosquitoes can be converted into fertile males by inserting just two genes on an autosome. Could be valuable for sex separation or gene drives to bias sex ratio. See Aryan et al. 2020 and Biedler et al. 2024.
Jeremy Herren (@JeremyHerren) shared work on microsporidia MB, a mosquito symbiont with potential to block malaria transmission. The @Symbio_Vector project has new data on how microsporidia infection affects mosquito gene expression and immunity. See Makhulu et al. 2023.
Nsa Dada spoke about the Anopheles mosquito microbiome and it’s potential role in insecticide resistance. Evidence that the microbiome differs between resistant and susceptible mosquitoes, replicated in two mosquito species. Is it causal? See Omoke et al. 2021 and Pelloquin et al. 2021.
Mary Ann McDowell spoke about population genetics and metagenomics of sandflies which transmit leishmania. Evidence for cryptic species in Brazil, associated with differences in male courtship song and microbiome. See Labbé et al. 2023.
Michael Branstetter (@bramic21) presented the Beenome100 project which is assembling genomes for bee species important for agriculture and conservation. See beenome100.org.
Matilda Gikonyo used trio binning to improve genome assemblies of small herbivorous insects with high genetic diversity, enabling resolution of maternal and paternal haplotypes.
Sarah Dyer presented the new rapid.ensembl.org site which releases new genomes every 2 weeks, keeping up with all the new genomes from tree of life and earth biogenome projects. Also the main @ensembl site has has a complete makeover, see beta.ensembl.org.
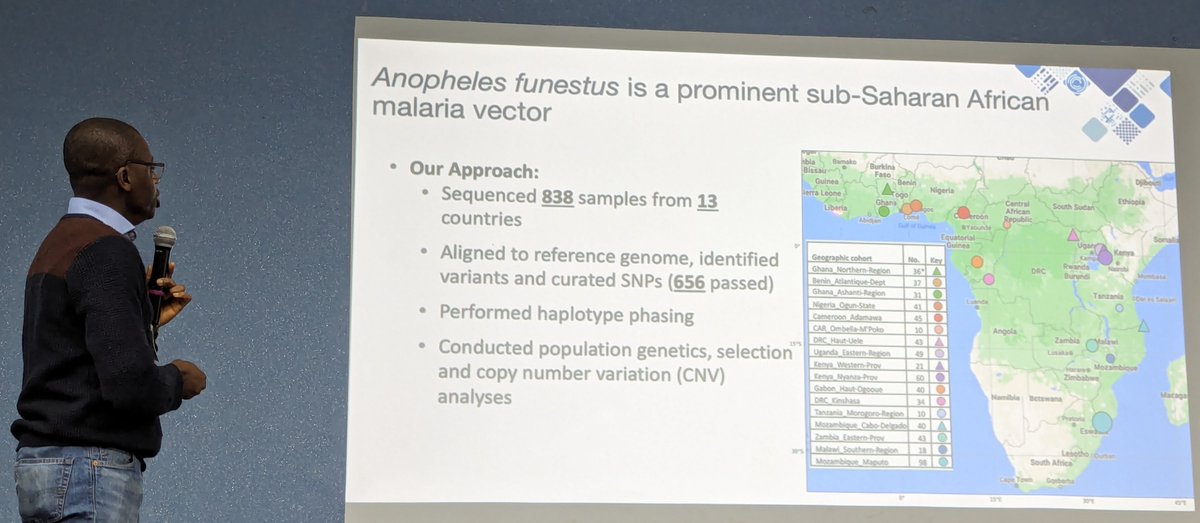
Joachim Nwezeobi presented on the population genomics of Anopheles funestus from across Africa. New results on population structure, chromosomal inversions, recent selection, copy number variation and adaptive gene flow. Paper coming soon from Joachim and @mariloubodde.
Tamar Carter is using population genomics to track the invasive malaria vector Anopheles stephensi. The spread of stephensi in Ethiopia seems to align with transport networks. Also stephensi in Yemen are structured into two distinct populations. See Carter et al. 2021, Samake et al. 2023 and Assada et al. 2024.
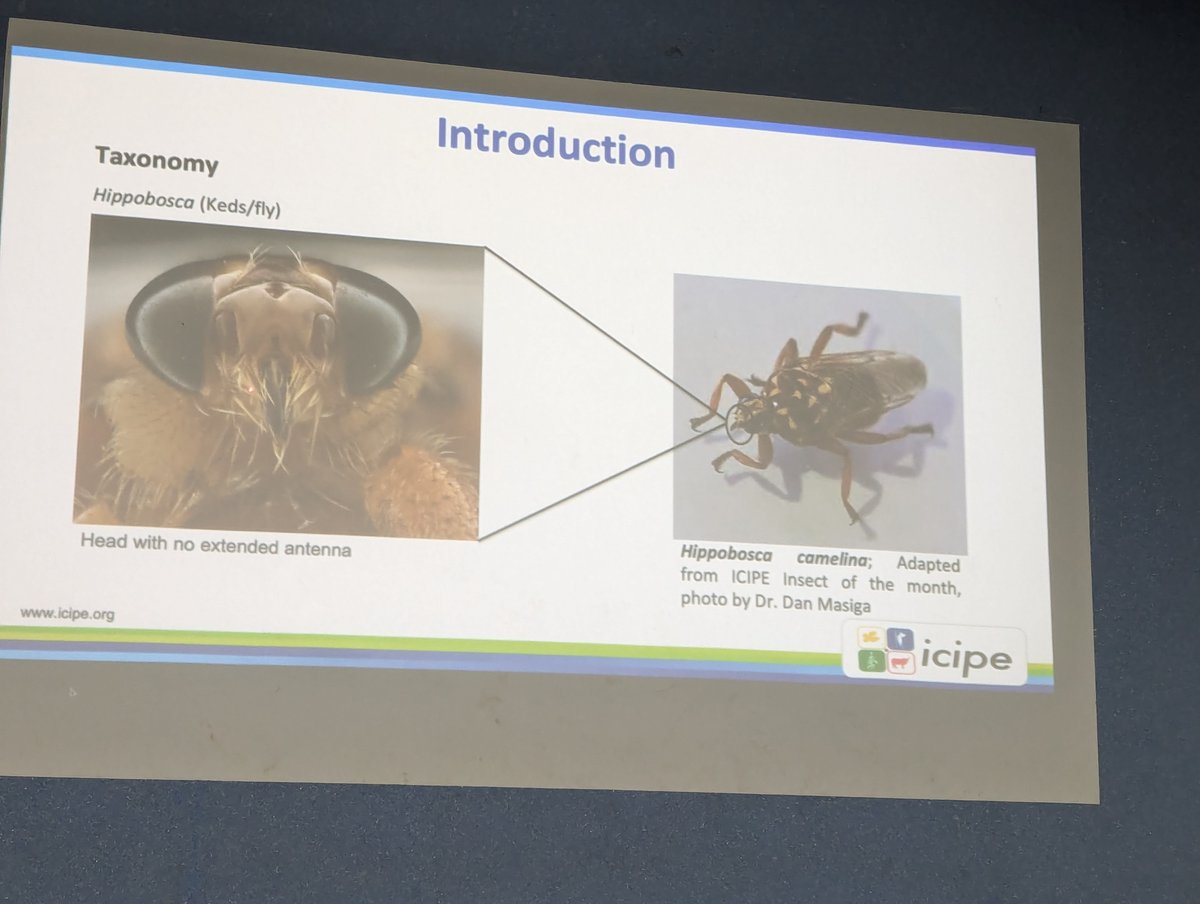
Fredrick Kebaso (@fredrickkebaso) assembled a genome for the camel ked, a biting fly that transmits various pathogens and affects the health of camels, making life difficult for marginalised communities in the Horn of Africa coping with climate change.
Billiah Bwana improved the annotation of genes in the tsetse fly expressed in antennae and involved in olfaction. Important because odour-baited traps are used for tsetse control. See Bwana et al. 2022.
Luna Kamau presented the discovery of Anopheles coluzzii in Kenya, which is genetically related to sahelian An. coluzzii from Mali and Burkina Faso and shares insecticide resistance alleles with these West African populations. See Kamau et al. 2024.
Megan Fritz (@MosquitoDoc) showed how the corn earworm has evolved resistance to transgenic (Bt) corn via copy number variation at a cluster of trypsin genes. Genomic monitoring could be used to detect resistance emerging in as little as 1 generation. See Pezzini et al. 2024 and Taylor et al. 2024.
Isaiah Debrah (@ISAIAHDEBRAH1) has found non-coding RNAs which are upregulated in insecticide resistant Anopheles funestus in western Kenya. The role of ncRNAs in regulating expression of detoxification genes needs more study. See Debrah et al. 2024.
Diana Omoke investigated gene expression profiles of Anopheles arabiensis populations from Western Kenya resistant to multiple insecticide classes. Resistant mosquitoes overexpressed salivary and cuticular proteins in addition to multiple detoxification genes. See Omoke et al. 2024.
Mahamat Gadji (@2017Kafir) reported genome-wide association analysis of Anopheles funestus from Mibellon in Cameroon highly resistant to pyrethroid insecticides. Strong selection at Cyp6 (rp1) and Cyp9 loci. See Gadji et al. 2024.
That’s it for the science. Going to be hard to beat this for a conference excursion, half a day at the Nairobi National Park…
Huge thanks to @SamRund, @Tung_Jowi, Mary Ann for organising a fantastic meeting, and to @pamcafrica, @KEMRI_Kenya, @ndeckinstitute, @Arthropod_i5k, @IGBIllinois, @kstatebio, @NIH, for making it possible.
Looking forward to the next AGS, wherever in the world it may be!